Research
Recent poster briefly describing our research and the lab. Below the poster, you will find a more detailed description of our main research areas.

Colon cancer
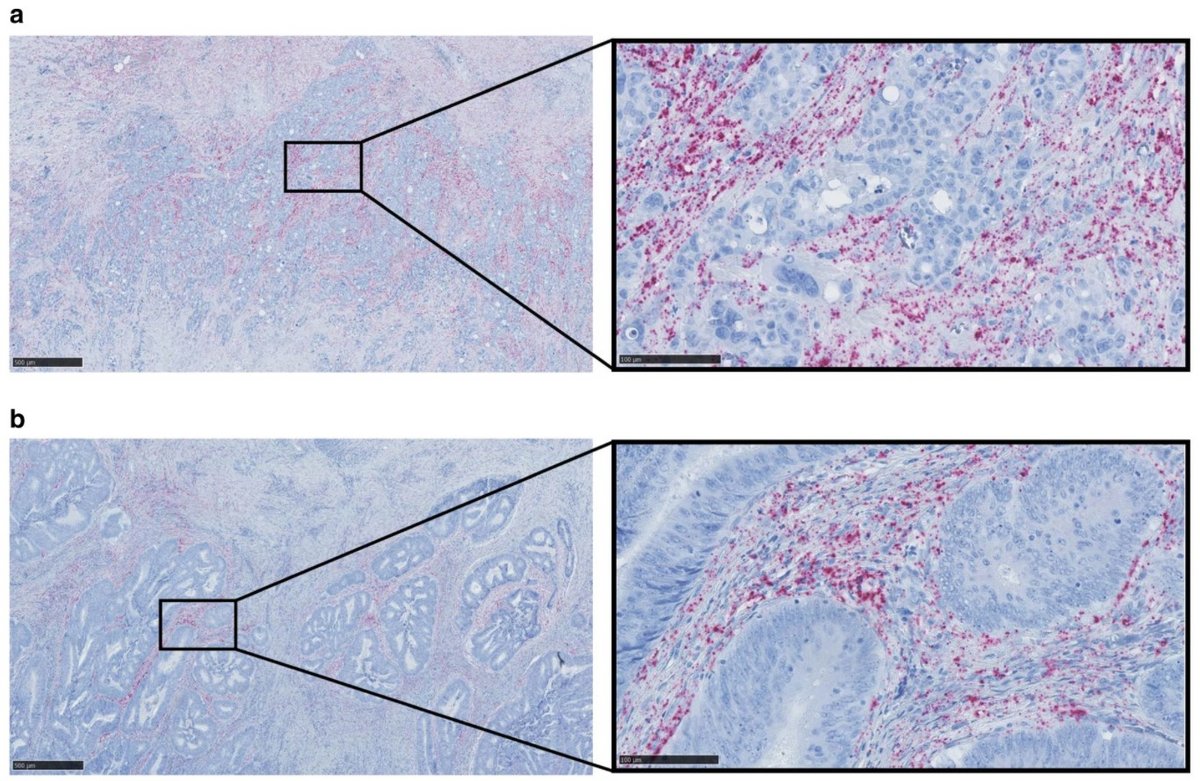
Colon cancer (CC) is one of the most common cancers in the western world and the high incidence is associated with the western lifestyle. At the time of diagnosis, approximately 70% of the patients have a loco-regional disease making them potentially curable by surgery and due to an aging population and better screening programs, more and more cancer patients are diagnosed at an earlier disease stage. However, the metastatic spreading of cancer cells to new parts of the body continues to be a very deadly aspect of CC, yet it is a poorly understood and complex process. Thus, we urgently need to get a better understanding of the metastatic process at the molecular level in order to enable the development of biomarkers that are capable of predicting the presence of occult metastases (undetectable micro-metastases) and to identify and validate targets for new treatments that specifically eliminate metastases.
We aim to make new ground-breaking discoveries in cancer biology that have translational potential using state-of-the-art technologies, including spatial transcriptomics. In addition, we focus on a newly discovered class of biomolecules, the circular RNAs (circRNAs), with strong gene-regulatory potential. These endogenous molecules are currently severely understudied, as they are discarded in most library preparation protocols for high-throughput RNA sequencing (RNA-seq) due to their lack of poly(A) tails. Thus, most publically available RNA-seq datasets, including the data from The Cancer Genome Atlas (TCGA), cannot be analyzed for circRNA expression. Therefore, circRNAs represent a hitherto overlooked resource of biomolecules with potential clinical value in cancer diagnostics, prognostics, and prediction of response to treatment. This is substantiated by circRNAs being detectable in body fluids, due to their covalently closed structure renders them extremely stable, allowing for non-invasive testing. Learn more about this project in the Open Discovery Innovation Network (ODIN) introductory video.
These projects are possible thanks to funding from the Novo Nordisk Foundation, the Danish Cancer Society, Riisfort Fonden and Magda Sofie og Aase Lütz's Mindelegat.
Links to publications:
The emerging roles of circRNAs in cancer and oncology, Nature Reviews Clinical Oncology, 2022
The biogenesis, biology and characterization of circular RNAs, Nature Reviews Genetics, 2019
Circular RNAs in cancer: opportunities and challenges in the field, Oncogene, 2018
Hematological malignancies
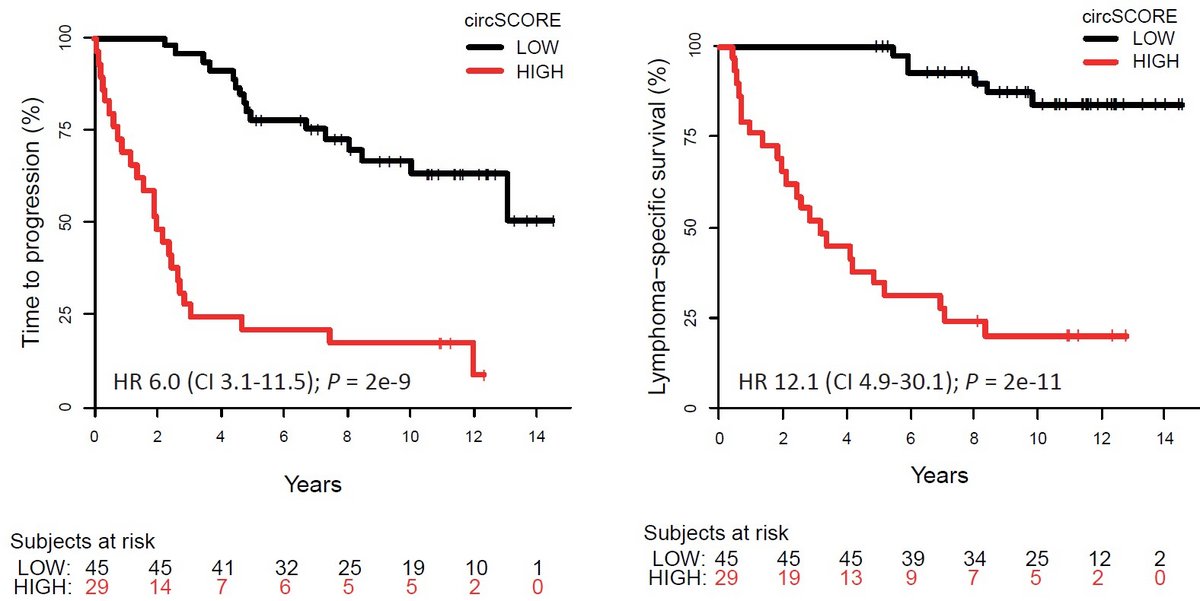
We study the molecular functions and clinical potential of circular RNAs (circRNAs) and other long non-coding RNAs (lncRNAs) in several hematological malignancies, including multiple myeloma, mantle cell lymphoma and myelodysplastic syndromes. Based on genome-wide expression profiling using RNA-sequencing, we find potential interesting circRNA and lncRNA candidates in these diseases. Our overall aim is to characterize of the (epi)genetic mechanisms behind aberrant circRNA expression in cancer and the molecular functions of circRNAs being drivers of cancer pathogenesis. In particular, we correlate changes in circRNA expression patterns to clinically relevant mutations in epigenetic regulators and spliceosome genes. In addition, we map genome-wide epigenetic changes in regulatory regions of circRNA host genes as well as the flanking regions of the splice sites involved in circRNA biogenesis. Finally, we aim to translate interesting findings into clinical use, by discovering individual circRNAs, or panels of circRNAs, which have diagnostic, prognostic or predictive value as well as to identify specific circRNAs, which can be used as targets for novel treatment strategies.
These projects are possible thanks to funding from the Lundbeck Foundation and Eva og Henry Frænkels Mindefond.
Links to publications:
Impact of U2AF1 mutations on circular RNA expression in myelodysplastic neoplasms, Leukemia, 2023
The emerging roles of circRNAs in cancer and oncology, Nature Reviews Clinical Oncology, 2022
The biogenesis, biology and characterization of circular RNAs, Nature Reviews Genetics, 2019
Circular RNAs in cancer: opportunities and challenges in the field, Oncogene, 2018
Pediatric Brain Tumors
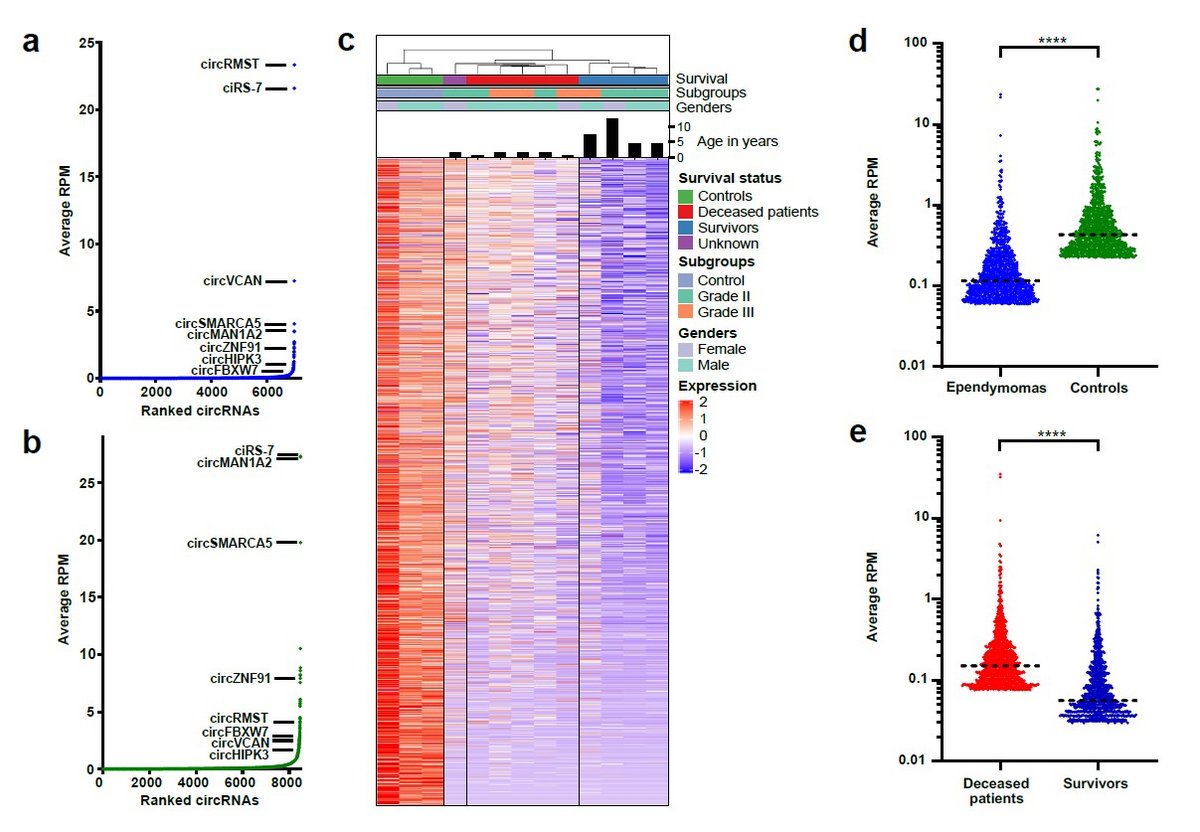
Pediatric brain tumors mainly develop in the cerebellum, with ependymoma, medulloblastoma and pilocytic astrocytoma as the most prevalent subtypes. Clinical management is challenging, and pediatric patients with intracranial ependymoma and medulloblastoma have high mortality rates. These tumors are currently treated using non-targeted therapies, because few somatically mutated driver genes are present and the underlying pathobiology is poorly described. Thus, in the majority of cases, the underlying oncogenic drivers are unknown and prediction of patient outcome based on tumor location, clinical characteristics and histopathology is challenging. Consequently, an improved understanding of the underlying pathobiology at the molecular level would provide much needed predictive power for patient outcome as well as new avenues for therapeutic intervention.
Circular RNAs (circRNAs) and other non-coding RNAs have recently emerged as key players in the pathobiology of many different human cancers. However, nothing is currently known about circRNAs Pediatric brain tumors, with the exception of our own recent data published in Brain Pathology. Thus, circRNAs represent a hitherto overlooked resource of biomolecules with potential value in cancer diagnostics, prognostics, and for prediction of response to treatment. We aim to undercover the molecular functions of key upregulated circRNAs identified in our previous work and further uncover the translational potential of circRNAs in pediatric brain tumors using well-defined, population-based cohorts.
This project is possible thanks to funding from Børnecancerfonden, The Novo Nordisk Foundation, Danish Cancer Society and the Dagmar Marshalls Fond.
Links to publications:
The emerging roles of circRNAs in cancer and oncology, Nature Reviews Clinical Oncology, 2022
Distinct circular RNA expression profiles in pediatric ependymomas, Brain Pathology, 2020
The biogenesis, biology and characterization of circular RNAs, Nature Reviews Genetics, 2019
Circular RNAs in cancer: opportunities and challenges in the field, Oncogene, 2018
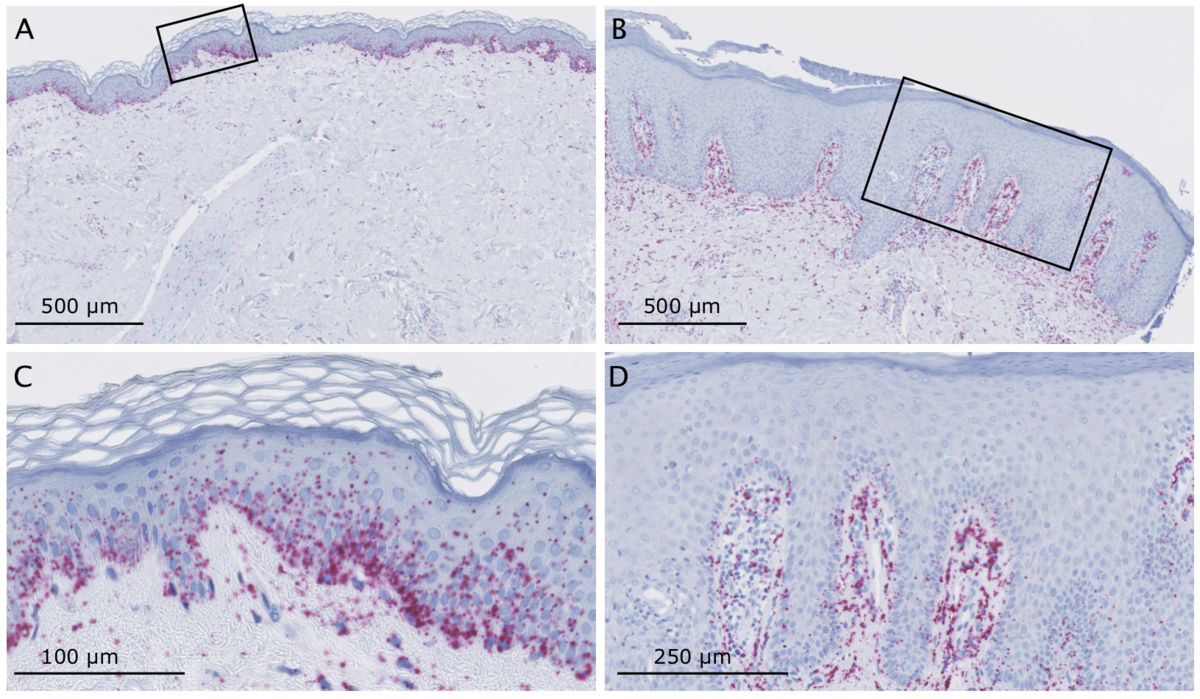
Inflammatory skin diseases
The skin is involved in the innate immune response as part of its protective function against pathogens and other environmental insults. Inflammation is one of the key responses of the innate immune system, which can be triggered by infection, injury and irritation, resulting in symptoms such as pain, heat, redness and swelling. The inflammation serve to eliminate the intruders as well as damaged cells. However, long-term and uncontrolled inflammation can lead to chronic cutaneous diseases such as psoriasis and atopic dermatitis, and chronic inflammation is one of the hallmarks of solar ultraviolet radiation and skin cancers mediated by other environmental agents. Inflammatory skin diseases affect a large proportion of the population and a better understanding of the molecular mechanisms underlying skin inflammation is crucial to develop better treatments and discover biomarkers for predicting response to existing treatments.
Circular RNAs (circRNAs) have recently emerged as an abundant class of non-coding RNAs with strong gene-regulatory potential. In particular, our collaborators have discovered a circRNA, named ciRS-7, with more than 70 highly conserved binding sites for miR-7, which we have recently shown to be extremely deregulated in a spatially defined manner in inflamed skin. However, ciRS-7 and circRNAs in general are currently severely understudied and virtually nothing is known about the mechanisms behind the aberrant expression of circRNAs and their function in inflammatory skin diseases.
This project is possible thanks to funding from the LEO Foundation.
Links to publications:
The biogenesis, biology and characterization of circular RNAs, Nature Reviews Genetics, 2019