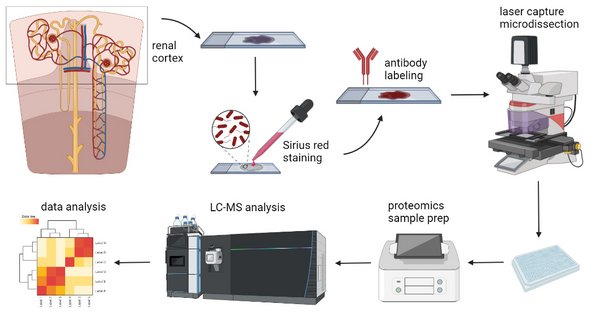
Research
The major current research themes are:
Systems biology of chronic kidney disease (CKD)
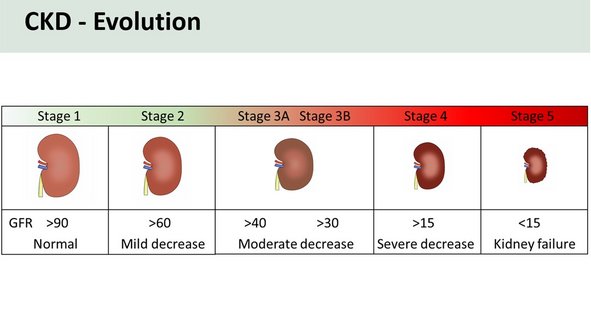
Chronic kidney disease (CKD) is one of the most deadly diseases faced by patients and is a major global health and socioeconomic burden. CKD increases cardiovascular morbidity and premature mortality and decreases quality of life. Hypertension (HTN) and type 2 diabetes mellitus (T2DM), which are reaching epidemic levels, are major risk factors for CKD.
Current strategies to diagnose CKD and its progression are suboptimal. Thus, there is an urgent need for new approaches for early detection of the most “at risk” individuals and identification of CKD signatures to aid in designing novel drugs and preventive measures that could ameliorate progression of CKD.
Previous research endeavors to identify molecular signatures that predict CKD progression have not been hugely successful, which may be due to the poor correlation of genetic information and cellular phenotypes, or the obstacle of not knowing which animal model of CKD reflects human pathophysiology on a molecular level.
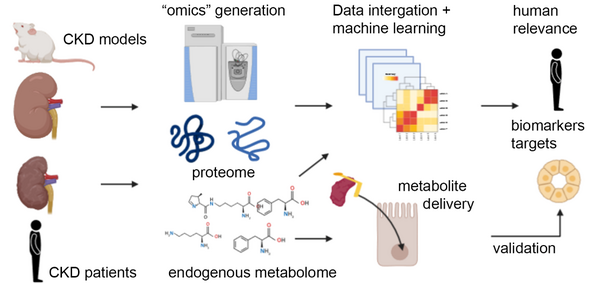
In this project, funded by the Open Discovery Innovation Network (ODIN) at Aarhus University alongside Markus Rinschen, Kenneth Howard, Ira Assent and researchers at Astra Zeneca, our overarching goal is to identify metabolites that predict kidney cell phenotypes during CKD and how crosstalk of these metabolites with the proteome drive CKD progression. We aim to identify common metabolites that alter kidney and cellular physiology and can act as diagnostic biomarkers and future therapeutic targets of CKD.
Potassium homeostasis and blood pressure
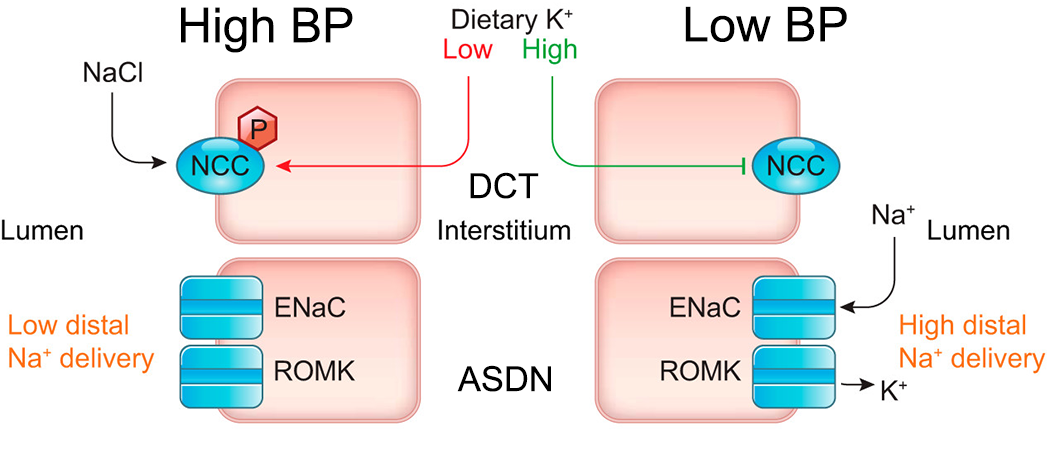
Hypertension (HTN) is the single greatest contributor to premature death and disability in the world. Sodium (Na+) rich diets are a major contributor to the onset of HTN, but efforts to reduce population Na+ intake have had modest success. Importantly, a low K+ intake accompanies the high Na+ intake in western diets. This may be an alternative avenue for intervention as there is strong experimental evidence linking K+ and BP. The mechanisms underlying the effects of K+, and its implications for prevention and treatment of HTN, remained obscure until our research contributed to the development of a new mechanism, defined the “renal K+ switch”. In this multidisciplinary research program utilizing gene editing, physiological models, proteomics, cell biology, high-res imaging, and human studies we aim to rapidly accelerate the understanding of this new pathway. The overarching objectives are to: 1) delineate the molecular, structural and physiological basis of the K+ switch; 2) understand factors that determine K+ sensitivity; 3) identify new targets for therapeutic and preventative intervention; 4) increase understanding of how dietary K+ helps maintain healthy BP for aiding public health and HTN prevention
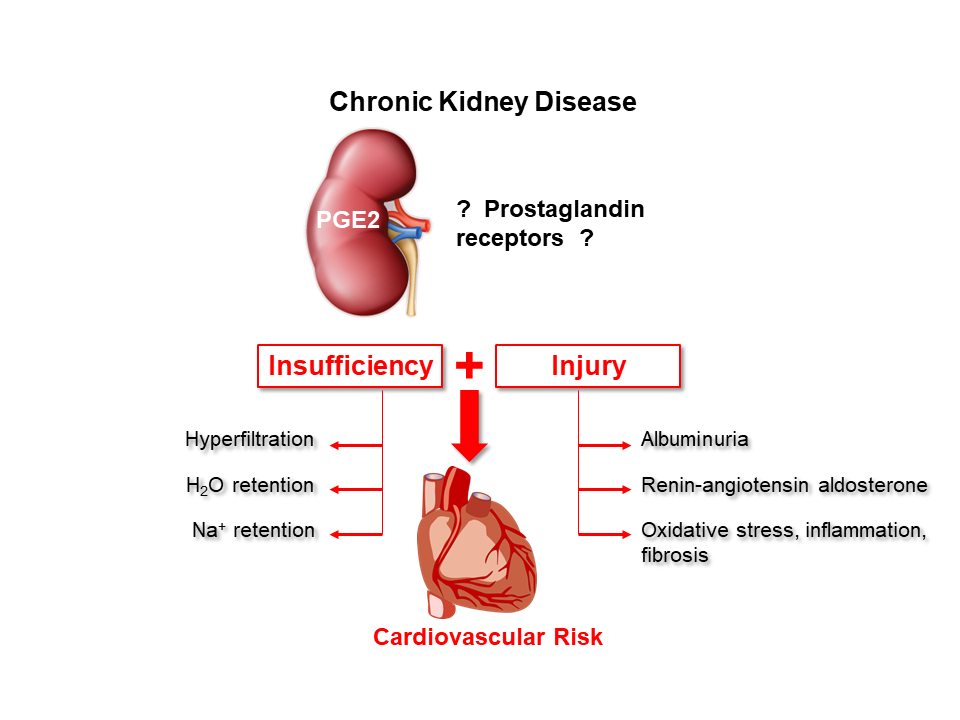
Prostaglandins in kidney physiology and pathophysiology
Prostaglandin E2 (PGE2), a product of membrane phospholipid metabolism, is a signaling molecule that acts via activation of one or more of the four PGE2 receptors (EP1-4). In the kidney, prostaglandins regulate vascular, glomerular, and tubular function, rendering them a unique target for chronic kidney disease (CKD) treatment - the pathogenesis of which involves interplay between all these components. CKD is a major global health burden with an urgent and unmet need for improved mechanistic insights. In this project we are examining the role of EP1-EP4 in various aspects of kidney physiology, but also determining the potential of modulating renal prostaglandin effects as a novel intervention for CKD. We are using a targeted assessment of prostaglandins effects during CKD in mice and at a clinical level using various human interventional cohorts. We hope that successful completion of this multidisciplinary research program will deepen our understanding of CKD pathogenesis, leading to clinical application and ultimately contribute to CKD prevention.
Single Cell Proteomics
The kidney is composed of multiple cell types that have unique functions and together contribute to the overall homeostatic role of the kidney. Knowledge of each of these different cell types, and their heterogeneity will provide insights into their specific functions, as well as provide a molecular platform for understanding the role of these cells in kidney disease. However, it is increasingly recognized that mRNA levels do not correlate with protein levels (and post-translational modifications) that critically determine cell behaviors and phenotypes. Therefore, advancing our understanding of individual cell function requires assessment of the proteome. Until recently, such large-scale studies on individual cells have been a fantasy for the field. However now, aided by developments in mass spectrometry (MS) that increase sensitivity by orders of magnitude, we are able to quantify thousands of proteins/proteoforms in individual cells/cell types and generate a molecular landscape of the heterogeneity of healthy and diseased kidney cells.